library(rgbif)
library(tidyverse)
library(data.table)
library(maps)Another species found! I am attempting to ID all the conifer species that occur in the Miracle Mile while out on runs so I can make a visit to the Miracle Mile and ID all of them in one go.

These sugar pine cones are huge! I have a few in my pinecone collection that are greater than 35 cm! Sugar pines (Pinus lambertiana) are fairly common species and I see them on trail runs often. For this post I first pulled the observations from GBIF, plotted them, and then looked where I had recently ran that would overlap with the species observation data. In this case, it was on a section of the Pacific Crest Trail (PCT) that I did an out and back run on.
Subset our search to only Northern California
mm_geometry <- paste('POLYGON((-124.4323 42.0021, -121.5045 42.0021, -121.5045 40.194, -124.4323 40.194, -124.4323 42.0021))')Pull in the publicly available data and make a few quick plots.
pinus_lambertiana_NC <- occ_data(scientificName = "Pinus lambertiana", hasCoordinate = TRUE, limit = 10000,
geometry = mm_geometry )
head(pinus_lambertiana_NC)$meta
$meta$offset
[1] 300
$meta$limit
[1] 2
$meta$endOfRecords
[1] TRUE
$meta$count
[1] 302
$data
# A tibble: 302 × 120
key scien…¹ decim…² decim…³ issues datas…⁴ publi…⁵ insta…⁶ publi…⁷ proto…⁸
<chr> <chr> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr>
1 40463… Pinus … 41.3 -122. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
2 40463… Pinus … 41.3 -122. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
3 34993… Pinus … 41.9 -124. cdc 50c950… 28eb1a… 997448… US DWC_AR…
4 37057… Pinus … 41.5 -124. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
5 37053… Pinus … 41.2 -122. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
6 37472… Pinus … 40.9 -124. cdc 50c950… 28eb1a… 997448… US DWC_AR…
7 37598… Pinus … 41.8 -124. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
8 39613… Pinus … 40.4 -123. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
9 37642… Pinus … 41.9 -124. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
10 37603… Pinus … 41.9 -124. cdc,c… 50c950… 28eb1a… 997448… US DWC_AR…
# … with 292 more rows, 110 more variables: lastCrawled <chr>,
# lastParsed <chr>, crawlId <int>, hostingOrganizationKey <chr>,
# basisOfRecord <chr>, occurrenceStatus <chr>, taxonKey <int>,
# kingdomKey <int>, phylumKey <int>, classKey <int>, orderKey <int>,
# familyKey <int>, genusKey <int>, speciesKey <int>, acceptedTaxonKey <int>,
# acceptedScientificName <chr>, kingdom <chr>, phylum <chr>, order <chr>,
# family <chr>, genus <chr>, species <chr>, genericName <chr>, …pinus_lambertiana_coords <- pinus_lambertiana_NC$data[ , c("decimalLongitude", "decimalLatitude", "occurrenceStatus", "coordinateUncertaintyInMeters", "institutionCode", "references")]
maps::map(database = "state", region = "california")
points(pinus_lambertiana_coords[ , c("decimalLongitude", "decimalLatitude")], pch = ".", col = "red", cex = 3)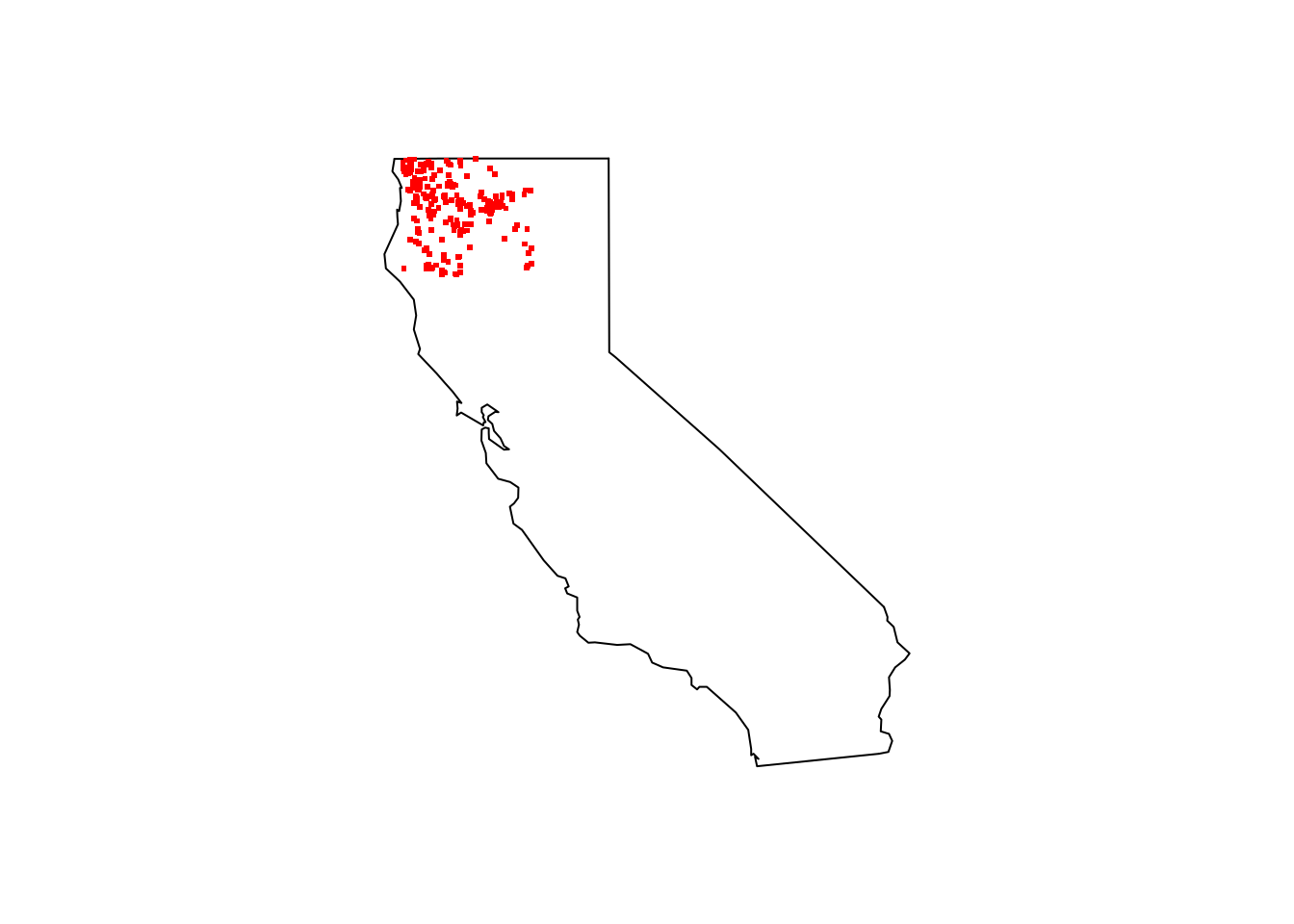
Pull in my run data.
TrailRun1 <- read.csv("~/DATA/data/TrailRun_SugarPine.csv")Make a quick plot.
TrailRun_plot <- ggplot(TrailRun1, aes(x = position_long, y = position_lat)) +
coord_quickmap() + geom_point() +
xlab("Longitude") + ylab("Latitude")
TrailRun_plot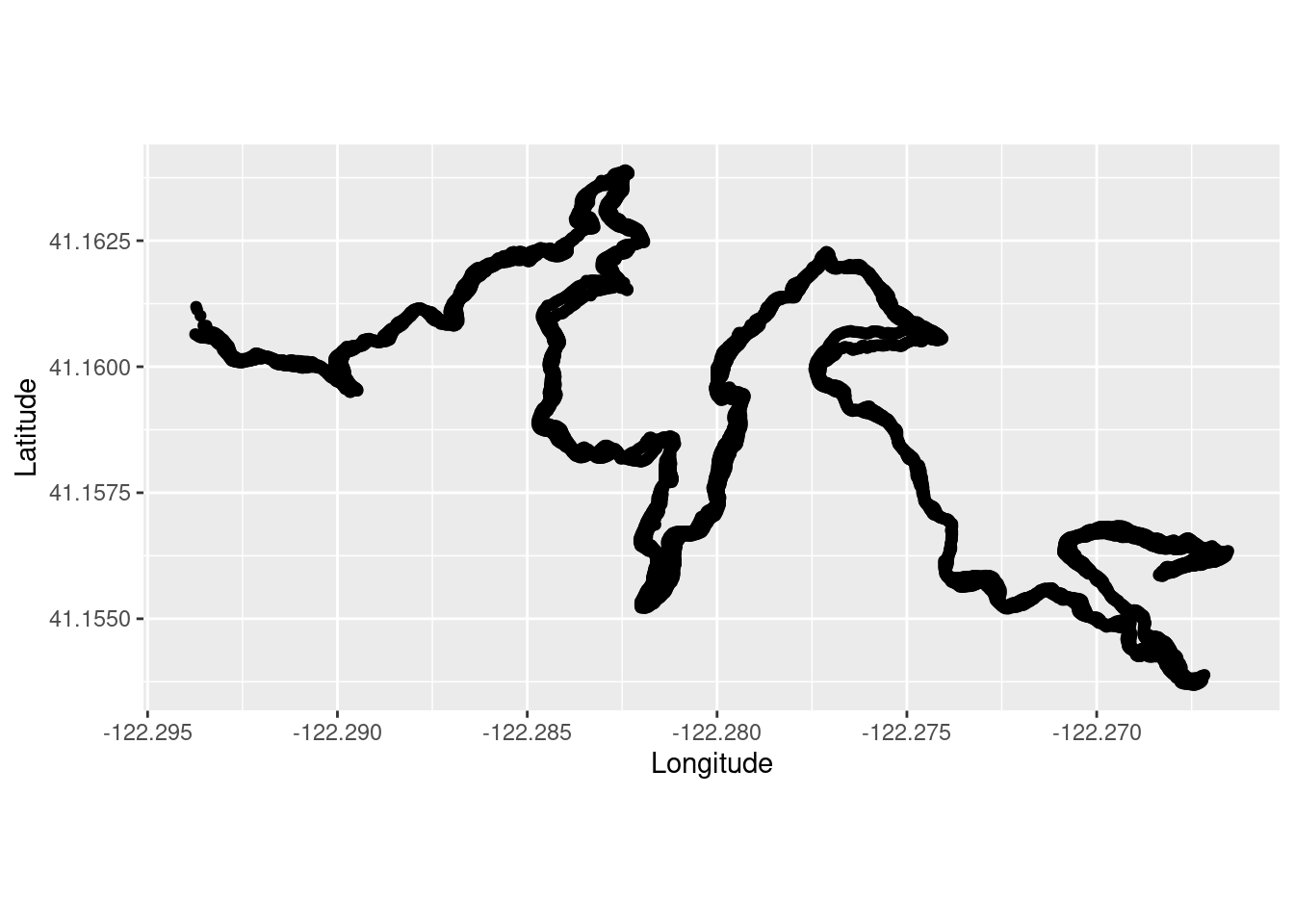
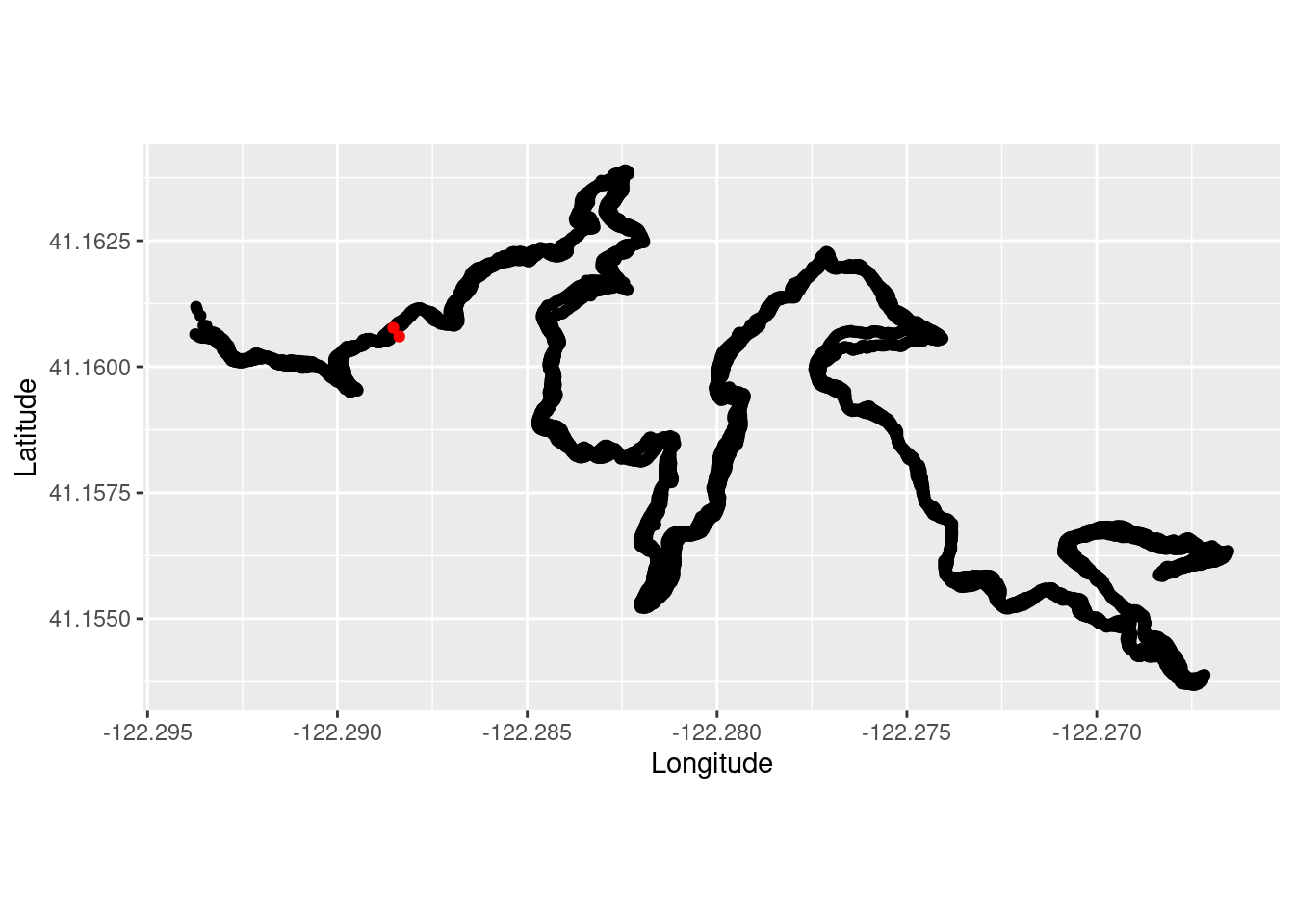
Subset the larger California dataset to only include a zoomed in portion where I ran based on the maximum and minimum coordinates from the run plot above.
pinus_lamb_coords_sub <- subset(pinus_lambertiana_coords, decimalLatitude > 41.1525 & decimalLatitude < 41.1650 & decimalLongitude > -122.295 & decimalLongitude < -122.265 )Overlay the plots!
TrailRun_plot2 <- ggplot() +
coord_quickmap() +
geom_point(data = TrailRun1, aes(x = position_long, y = position_lat), color = 'black') +
geom_point(data=pinus_lamb_coords_sub, aes(x = decimalLongitude, y = decimalLatitude), color = 'red') +
xlab("Longitude") + ylab("Latitude")
TrailRun_plot2